Research
The Kwapis Lab studies the molecular and epigenetic mechanisms that underlie long-term memory formation, storage, and updating. We want to understand the mechanisms that underlie memory and identify how these mechanisms are dysregulated during aging, leading to age-related memory impairments.
Our research uses cutting-edge techniques to dissect the molecular mechanisms that underlie memory and age-related memory decline. These techniques include:
-in vivo CRISPRi and CRISPRa to reduce or drive transcription in a site- and circuit-specific manner
-Modified CRISPR/dCas9 systems to remodel chromatin at selective genes
-Viral vectors in combination with cloning to manipulate gene expression or activity
-CatFISH to identify active neuronal ensembles
-Sophisticated behavioral techniques include fear conditioning, object location memory, and memory updating paradigms.
Current Projects
Age-related epigenetic repression of Per1 could have different outcomes depending on the brain region affected.
Understand how epigenetic regulation of the circadian clock gene Per1 contributes to age-related impairments in long-term memory
Aging is accompanied by impairments in both long-term memory and circadian rhythmicity. Although it is well-documented that circadian rhythms can influence memory formation, it is unclear how age-related deficits in these biological processes are related. Our work has shown that a key circadian gene, Period 1 (Per1) is repressed by the histone deacetylase HDAC3 in the aging hippocampus, leading to age-related impairments in memory formation. Per1 may function to relay time-of-day information to memory processes, possibly serving to "gate" when memory can form across the circadian day/night cycle.
Future work in the lab will focus on understanding how Per1 modulates memory formation in the young and aging hippocampus. Using a combination of site-specific viral manipulation, co-IP followed by mass spectrometry, ChIP, and next-generation sequencing, we hope to better understand how Per1 and other circadian clock genes influence memory formation and contribute to age-related cognitive decline.
DIrecting site-specific chromatin modifications in memory consolidation
Numerous studies have demonstrated that chromatin modifying enzymes, like histone acetyltransferases (HATs) and histone deacetylases (HDACs). play an important role in long-term memory. In previous work, for example, we have shown that the repressive histone deacetylase HDAC3 negatively regulates memory for fear conditioning in the hippocampus and amygdala. To date, most of the work in this field has manipulated chromatin modifying enzymes across the genome, making it difficult to pinpoint the specific genes at which these chromatin modifications affect memory formation. To address this weakness, we have developed modified CRISPR/dCas9 systems to target chromatin modifying enzymes directly to specific genes, allowing us to test whether driving chromatin modifications at specific genes can affect long-term memory formation in the young and aging brain. This tool can be readily modified to target a range of genes in numerous brain areas following a wide variety of behaviors. Current work in our lab is testing whether targeting HDAC3 or CBP (an opposing HAT) to the Per1 promoter can modulate memory formation. Future research will extend this work to other circadian genes using additional chromatin modifying enzymes.
CRISPR system to drive site-specific chromatin remodeling.
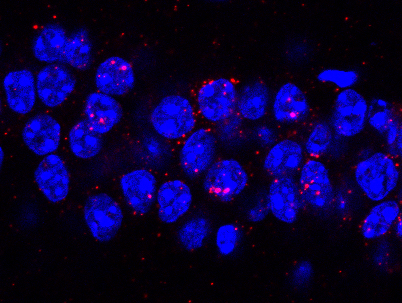
CatFISH used to identify active neuronal populations during memory updating.
Investigate the molecular and circuit-level mechanisms that support memory updating
Memory is not permanently stored in a fixed state but, rather, can be modified in response to new, relevant information. Stable memory undergoes a period of vulnerability following a retrieval session, a process termed "reconsolidation," during which the memory may be temporarily prepared to incorporate new information. Our work has demonstrated that reconsolidation-based updating procedures can be used to change the neural circuit supporting a fear memory. Further, we have developed a novel memory updating task that can assess the strength of both the original memory and the updated information in a single test session. Using this task, we have shown that updating engages an overlapping but distinct population of neurons in the dorsal hippocampus compared to the original memory. Further, aging mice show impairments in memory updating.
Our work aims to understand how memory is modified during updating. Using a combination of next-generation sequencing, ensemble-specific cell tagging, and DREADD-mediated manipulation, we hope to answer key questions about memory updating, including: are there distinct molecular mechanisms involved in memory updating that are not engaged by de novo memory formation? Can activating or inactivating the population of neurons engaged by the update session affect the original memory? Why is memory updating impaired in the aging brain?